The neXtProt SPARQL endpoint is accessible at: https://sparql.nextprot.org via HTTP GET or POST (see below).
We recommend our new users to start looking at our examples queries together with our data model.
- NXQ_00001 - Proteins phosphorylated and located in the cytoplasm
- NXQ_00004 - Proteins highly expressed in brain but not expressed in testis
- NXQ_00008 - Proteins whose genes are less than 50000 bp away from the location of the gene coding for protein p53
- NXQ_00147 - Proteins with a variant having an impact on the nucleus localization at level GOLD
- NXQ_00124 - What are the 10 most frequent families with member count
- NXQ_00126 - Peptides that are potential neo N-termini from undescribed isoforms
- NXQ_00148 - Variants with normal "ubiquitin-protein transferase activity" and decreased or increased binding to UBE2D1 (example: BRCA1-p.Ile89Thr)
Note that your saved queries can also be retrieved in the SNORQL environment if you are logged in.
Further links: You may want to have a look also at the presentation Practical Introduction to SPARQL and play with the The SPARQL playground.
SPARQL query with HTTP POST and CURL
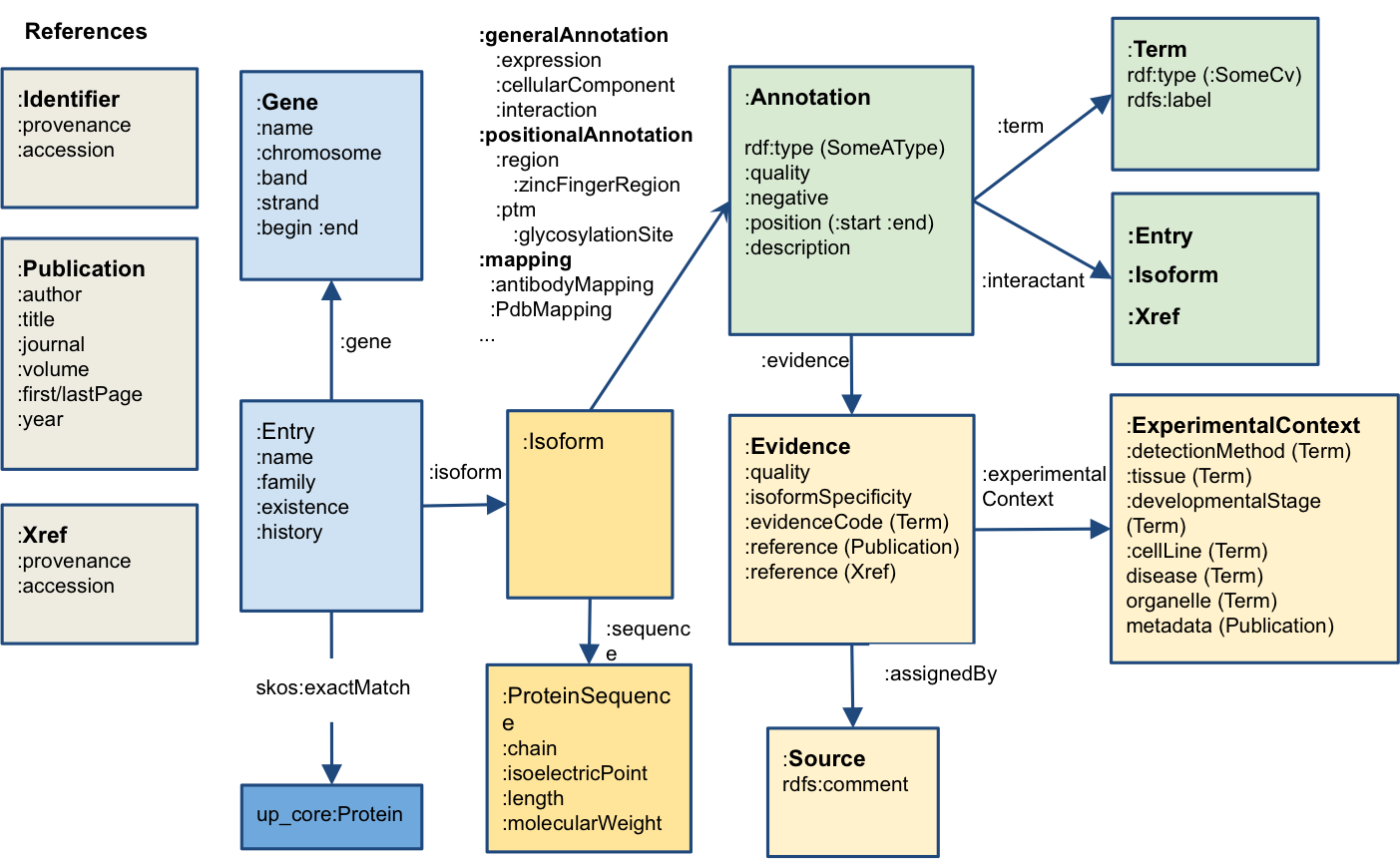
The following example, shows how to query the SPARQL endpoint using your UNIX terminal and saving your query in a file. Create a file called query.sparql and paste the following content:PREFIX : <http://nextprot.org/rdf#>
PREFIX cv: <http://nextprot.org/rdf/terminology/>
select distinct ?entry where {
?entry :isoform ?iso.
?iso :keyword / :term cv:KW-0597.
?iso :cellularComponent /:term /:childOf cv:SL-0086.
}QUERY=$(<query.sparql) && curl -X POST -H "Accept:application/sparql-results+json" --data-urlencode "query=$QUERY" https://sparql.nextprot.org/ You will find the complete list of prefixes in https://snorql.nextprot.org/ (click on PREFIX....)